Pioneering Anti-Inflammatory Therapy: The Path to Precision Therapy with C3aR Peptide Libraries
Abstract:
Bibliography/Citations:
Bibliography
Afshar-Kharghan, V. (2017). The role of the complement system in cancer. The Journal of clinical investigation, 127(3), 780-789.
Corcoran, J. A., & Napier, B. A. (2022). C3aR plays both sides in regulating resistance to bacterial infections. PLoS Pathogens, 18(8), e1010657.
Dalakas, M. C., Alexopoulos, H., & Spaeth, P. J. (2020). Complement in neurological disorders and emerging complement-targeted therapeutics. Nature Reviews Neurology, 16(11), 601-617.
Espinosa-Riquer, Z. P., Segura-Villalobos, D., Ramírez-Moreno, I. G., Pérez Rodríguez, M. J., Lamas, M., & Gonzalez-Espinosa, C. (2020). Signal transduction pathways activated by innate immunity in mast cells: translating sensing of changes into specific responses. Cells, 9(11), 2411.
Halai, R., Bellows-Peterson, M. L., Branchett, W., Smadbeck, J., Kieslich, C. A., Croker, D. E., Cooper, M. A., Morikis, D., Woodruff, T. M., & Floudas, C. A. (2014). Derivation of ligands for the complement C3a receptor from the C-terminus of C5a. European Journal of Pharmacology, 745, 176-181.
Hawksworth, O. A., Li, X. X., Coulthard, L. G., Wolvetang, E. J., & Woodruff, T. M. (2017). New concepts on the therapeutic control of complement anaphylatoxin receptors. Molecular immunology, 89, 36-43.
Janeway, C., Travers, P., Walport, M., & Shlomchik, M. (2001). Immunobiology: the immune system in health and disease (Vol. 2). Garland Pub. New York.
Ricklin, D., & Lambris, J. D. (2013). Complement in immune and inflammatory disorders: pathophysiological mechanisms. The Journal of Immunology, 190(8), 3831-3838.
Wang, Y., Zhang, H., & He, Y.-W. (2019). The complement receptors C3aR and C5aR are a new class of immune checkpoint receptor in cancer immunotherapy. Frontiers in immunology, 10, 439560.
Wende, E., Laudeley, R., Bleich, A., Bleich, E., Wetsel, R. A., Glage, S., & Klos, A. (2013). The complement anaphylatoxin C3a receptor (C3aR) contributes to the inflammatory response in dextran sulfate sodium (DSS)-induced colitis in mice. PLoS One, 8(4), e62257.
Yadav, M. K., Maharana, J., Yadav, R., Saha, S., Sarma, P., Soni, C., Singh, V., Saha, S., Ganguly, M., & Li, X. X. (2023). Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors. Cell, 186(22), 4956-4973. e4921.
Yadav, M. K., Yadav, R., Sarma, P., Maharana, J., Soni, C., Saha, S., Singh, V., Ganguly, M., Saha, S., & Khant, H. A. (2023). Structural insights into agonist-binding and activation of the human complement C3a receptor. bioRxiv, 2023.2002. 2009.527835.
Yan, Y., Tao, H., He, J., & Huang, S.-Y. (2020). The HDOCK server for integrated protein–protein docking. Nature protocols, 15(5), 1829-1852.
Zhang, Y.-y., & Ning, B.-t. (2021). Signaling pathways and intervention therapies in sepsis. Signal Transduction and Targeted Therapy, 6(1), 407.
Additional Project Information
Project files
Research Plan:
Pioneering Anti-Inflammatory Therapy: The Path to Precision Therapy with C3aR Peptide Libraries
Question Addressed
- C3aR/C3a function is partially important in the immune/complement system.
- Only a limited amount of research has been conducted on C3aR/C3a, leaving ample room for further investigation and exploration
- Recently the structure of the C3aR G-coupled receptor and the ligand C3a came out and was published on the protein structure database PDB (Protein Docking Bank)
- The C3aR/C3a receptor and ligand pair = chemotactic mediator in the human immune/complement system
- chemotactic mediator: helps mediate tissue damage and other inflammatory processes
- C3aR is dual-faceted as it can act as both an anti-inflammatory or proinflammatory receptor
- -> reason why designing a peptide that would act as an antagonist or agonist that would have a more potent and stronger binding and act better for this ligand and receptor pair to carry out its function is so important
- Could potentially be used to cure many diseases that involve inflammation in the human immune system
Goal
- Find out which mutations to which residues and what amino acid to change in the C3a ligand/protein (to help C3a better perform its function as an antagonist/agonist to the C3aR receptor)
- -> Design an ideal C3a molecule to help regulate inflammation in the complement/immune system
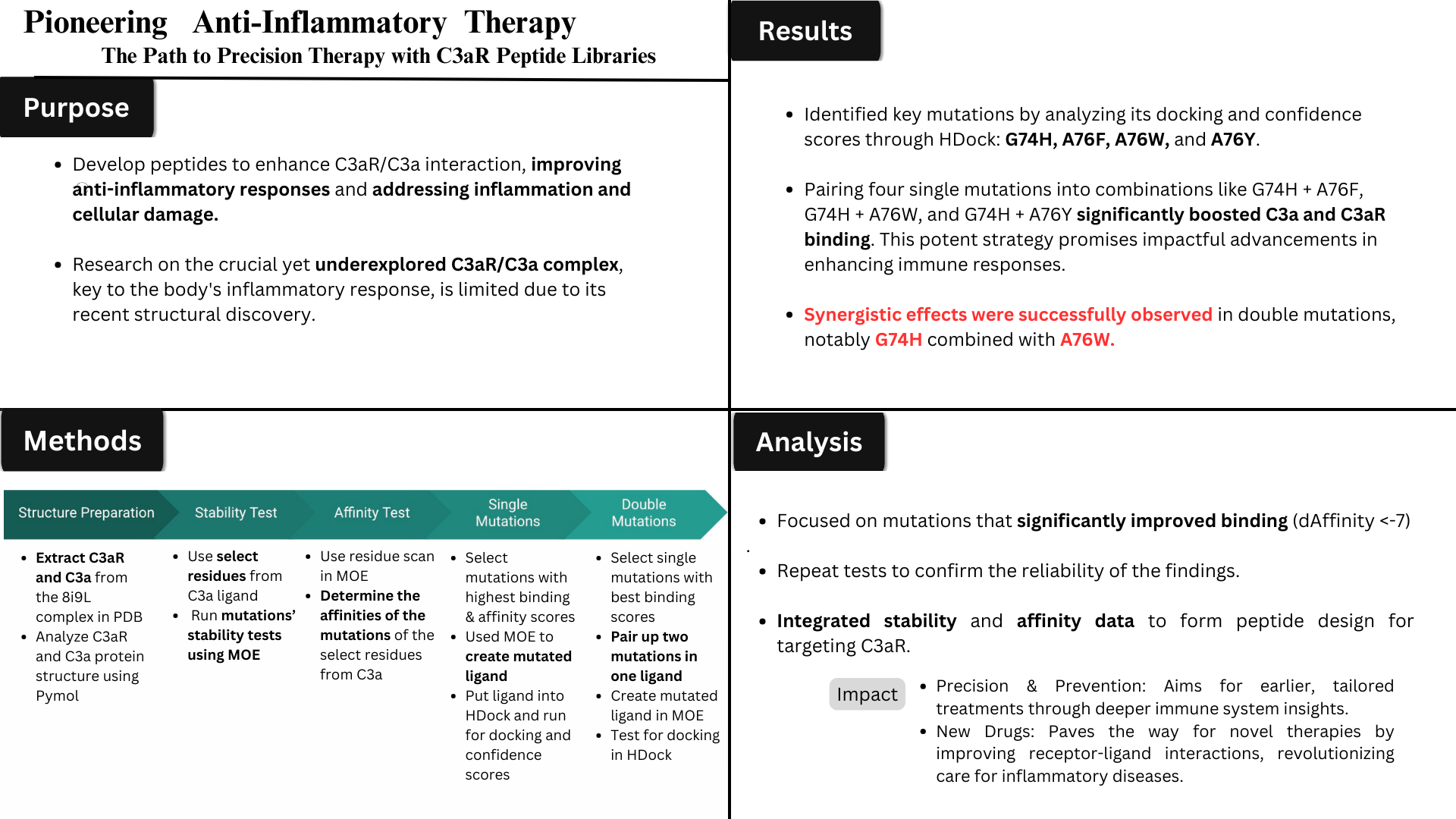
Method/Procedures
Procedures:
- Look at the recently published structure of the C3aR receptor and C3a ligand in Pymol (protein visualization software; models the protein’s structure)
- See how ligand and receptor bind together + what residues are involved in the active site
- HDock Server: used to visualize a model of C3a and C3aR binding together + get a general binding score and confidence score between these two proteins
- Figure out how well they bind together
- Also in HDock: a section that shows the particular strength of bonds between certain pairings of residues from each protein; this helps determine which residues need to be focused on to simulate mutations at a later stage
- Simulation of mutation(s)
- I will utilize the MOE (Molecular Operating Environment) software to carry out the simulations
- After that, I will mutate all residues that have been determined to be important in the process of the binding and active site
- After that, I will run the simulations of the residues being changed to each of the 19 other amino acids
- Results: MOE Software would provide the affinity and dAffinitiy scores of each of those mutations
- Upon analysis: these scores will help me see which mutations are potentially important and would help enhance the binding between the C3a ligand and C3aR receptor
Data Analysis:
Two main software used to conduct these trials and runs
- HDock Server
- I will simulate the binding between the ligand and receptor file being inputted and will use its pre-existing algorithm to give a visualization of various models it has predicted to be possible models for the binding between the C3aR receptor and ligand and all the confidence and docking scores
- This will show me which model is the best from all the models predicted by the algorithm; also shows the important bonds and strength of the bonds
- I will simulate the binding between the ligand and receptor file being inputted and will use its pre-existing algorithm to give a visualization of various models it has predicted to be possible models for the binding between the C3aR receptor and ligand and all the confidence and docking scores
- MOE (Molecular Operating Environment)
- Where I will run all the mutation stimulations
- It will generate affinity and dAffinity scores
- Affinity scores: how strong the bonds are
- dAffinity scores: how different the mutated residues’ affinity would be compared to the original and naturally occurring residue (the wildtype)
- Data analysis: It will give me a basis to determine if the mutation should even occur or not and help me determine which mutations of which residues are important
- ***It is by running multiple trials of mutation simulation trials that I will be able to determine which residues in the C3a ligand should be changed to what residues and amino acid
- ***Visual analysis of the mutation: use Pymol software
- shows the distances of those bonds and creates those bonds through computational methods
Risk and Safety
This project involves purely computer-based simulations, eliminating any potential risks or safety concerns.
Questions and Answers
Initial project questions
1. What was the major objective of your project and what was your plan to achieve it?
a. Was that goal the result of any specific situation, experience, or problem you encountered?
b. Were you trying to solve a problem, answer a question, or test a hypothesis?
The objective of my project was to try to design a peptide that would potentially enhance the binding and create an antagonist/agonist that was stronger than the current C3a protein itself to bind with the G-coupled receptor C3aR. The purpose of designing this was to enhance the function of the receptor C3aR in your body as this receptor controls inflammation in your body. Additionally, the C3aR/C3a function is partially important in the immune/complement system as it acts as a chemotactic mediator and is a dual-pathway meaning it can perform anti-inflammatory and pro-inflammatory functions depending on the cells and diseases. So I would say that I was trying to solve a problem and promote what could potentially be a stronger and more potent solution to certain diseases etc. in one’s body.
2. What were the major tasks you had to perform in order to complete your project?
a. For teams, describe what each member worked on.
For the design of the peptide, most of the work was done on protein and docking modeling software that uses the program/software’s algorithm to do so. In conclusion, most of the work was done on running trials on the computer and the software used rather than a laboratory. So the key component of the procedures and methods used for the experiments and the design process was just to run multiple trials determining the mutations’ affinities and dAffinities to determine the quality of each mutation’s dockings. Also,, I used other software to get the docking score and confidence scores as the software was able to model what the binding between the C3a and C3aR receptor would be and look like. Pymol is also a software that I used to be able to visualize how those proteins would look like and it was also the software that came in handy when analyzing the mutated ligands. It helped to create those bonds that stemmed from the mutated residue and also provided details about the bonds, for example how long it was and the angstroms involved with the bond. In total, 247 mutations and peptides were created throughout the whole research project but through various steps, the 247 mutations were narrowed down to 4 mutations that we focused on and used in our conclusion.
3. What is new or novel about your project?
a. Is there some aspect of your project's objective, or how you achieved it that you haven't done before?
b. Is your project's objective, or the way you implemented it, different from anything you have seen?
c. If you believe your work to be unique in some way, what research have you done to confirm that it is?
This is something that hasn’t been done yet as the complete structure and the chains and residues included in the C3aR receptor and C3a ligand had just been finalized and published recently so this kind of peptide designing for this particular pair of receptor and ligand has not been conducted yet. This would also pose a breakthrough as this is a newly published structure so being able to enhance its function and design its antagonist and agonist would be great for future uses in designing cures for certain diseases.
4. What was the most challenging part of completing your project?
a. What problems did you encounter, and how did you overcome them?
b. What did you learn from overcoming these problems?
Some of the problems I encountered were in being able to attain certain precision in the data across the trials of the mutations. One of the ways I mitigated this problem was by checking that all the methods used in all of the trials were similar. In MOE there are a lot of details that you need to make sure are in unison across all trials for the algorithm to perform the same job in all of the trials.
5. If you were going to do this project again, are there any things you would you do differently the next time?
One thing I noticed was that even though I was running the same mutations using the same software and thus the same algorithm, the affinities, and dAffinities for each of the times I ran it was different meaning that there probably is some sort of variability or uncertainty in the algorithm used in the software. So if I were to do this project again, I would probably just run more trials.
6. Did working on this project give you any ideas for other projects?
I think this project in the future if it were to be expanded on or continued, there could be future research conducted to determine whether more could be done. For example, using the design of the antagonist and agonist to create something that could pose as a cure or alleviate certain kinds of diseases that would attack the immune system. The C3aR/C3a receptor and ligand are used to function as either a pro-inflammatory or anti-inflammatory function in the immune/complement system.
7. How did COVID-19 affect the completion of your project?
COVID-19 restricted the usage of laboratories so it was harder for me to bring my research to the laboratory where I could’ve had access to technologies such as the Cryo-EM technology which would give me a direct actual visual of the protein complexes and possibly give me more accurate findings.